Pathway analysis in Strand NGS enables functional analysis of entities to understand their role in a biological process. The workflows in Strand NGS help identify genes which are significant in your experiment. These genes and data associated with them can be overlaid on pathways. Strand NGS provides direct access to curated pathway sources like WikiPathways and BioCyc. It is also packaged with a proprietary NLP derived interaction database from which Literature Derived Networks can be constructed.
Curated Pathways
Strand NGS can directly download the latest pathways from the WikiPathways website. We also host the BioCyc pathways on the Strand NGS server - these can be downloaded via the Annotation Manager. In addition, users can import any BioPAX (Level 2 or Level 3), or GPML files into the software.
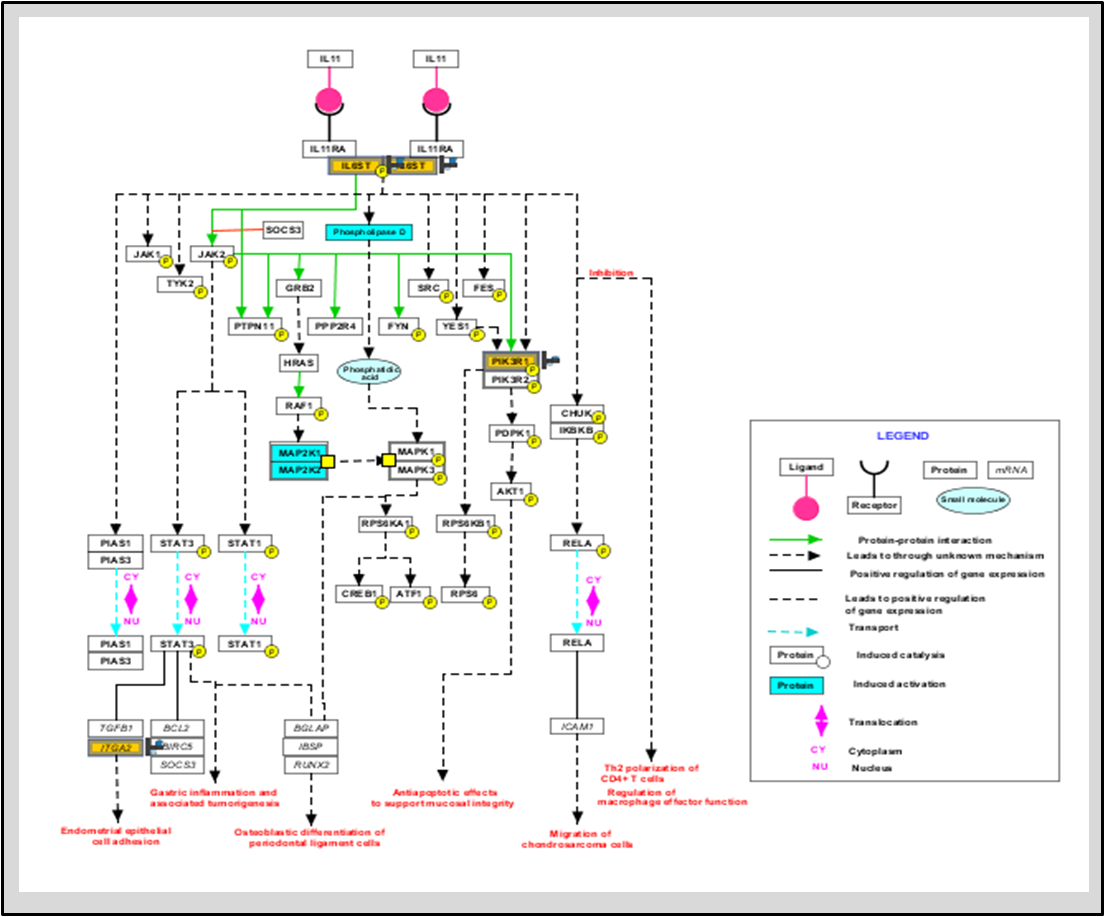
Interpret your experimental data through the following pathway analysis features in Strand NGS:
Single Experiment Analysis (SEA) compares an entity list with pathways in a pathway collection, identifies the common entities and computes the statistical significance of the pathway. The resulting pathway list can be filtered on the basis of number of common entities or the p-value to short list the most relevant pathways.
Multi-Omic Pathway Analysis (MOA) is similar to SEA but allows the user to choose entity lists from two different experiments. Entity list data from both the experiments can be superimposed on the pathway allowing users to get a better understanding of the role of the pathway in the two experiments.
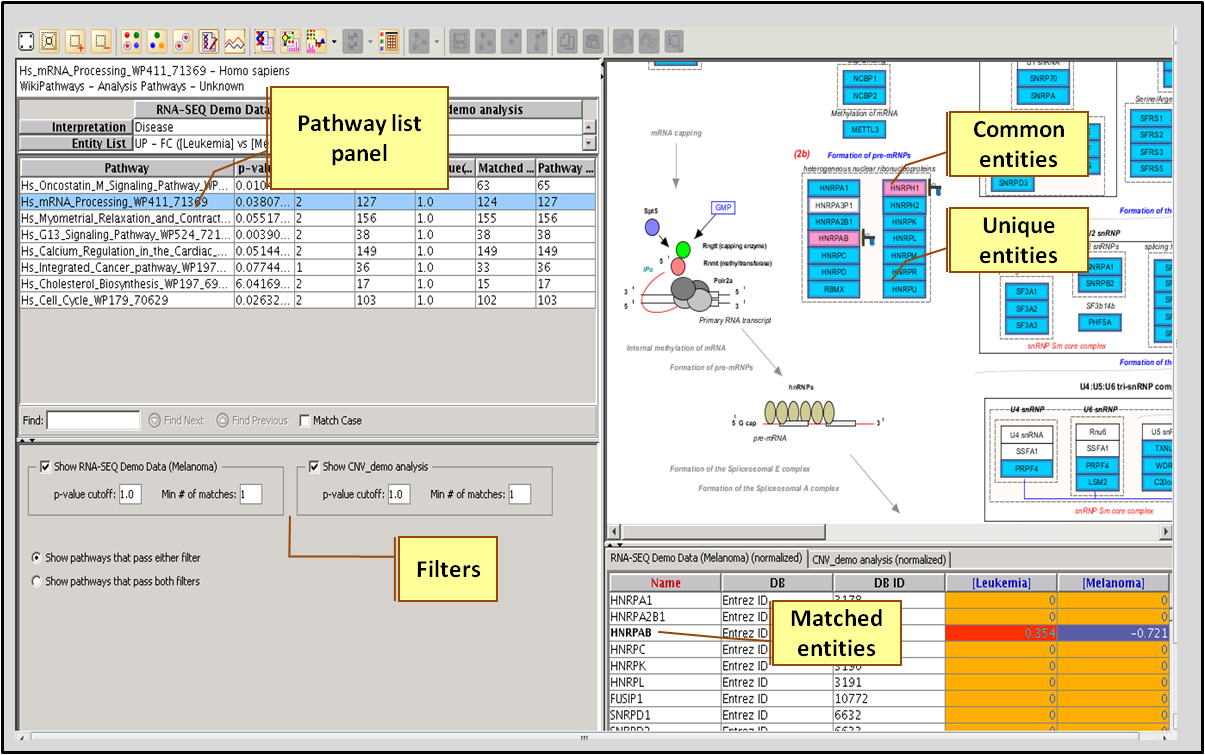
Choose a p-value cut-off and the minimum number of matched entities.
NLP Networks
NLP Network Discovery
An interaction database with millions of relations obtained by processing Medline content can be downloaded from the Annotation manager. Given a list of entities, you can query this database in a variety of ways to identify interactions involving the entities. These interactions can be used to build custom pathways.
Extract Relations Via NLP
Natural Language Processing (NLP) can be used to create new pathways from PubMed abstracts or full length publications. NLP first recognizes entities in sentences and then performs information extraction to identify relationships between these entities. Launch this NLP functionality by selecting NLP Networks => Extract Relations Via NLP from the Workflow.
MeSH Network Builder
The MeSH Network Builder creates pathways around a Term or a Concept. It obtains all the interactions containing these MeSH terms from the Interaction Database packaged with Strand NGS. Launch this NLP functionality by selecting NLP Networks => MeSH Network Builder from the Workflow.
GO analysis on an entity list identifies genes that have an over-representation of one or more GO terms that pass the cut-off p-value.
GSEA uses the ranked gene list to identify the gene sets that are significantly differentially expressed between two conditions.
GSA uses the S-Score, computed as a cumulative sum of t-Scores across the phenotypes for genes in a gene set for significance testing.
GSEA and GSA are available in the RNA-Seq experiments and require you to import Gene Sets before starting the analysis. Strand NGS supports the import of MIT-Harvard-Broad and custom gene sets in txt/csv, xml, grp or gmt file formats.
Download the Application Note on Integrated mRNA and microRNA transcriptome analysis Download the Data Import, Alignment, and Quality Control Guide Download the Application Note on Integrative RNA and ChIP-Seq analysis of regulatory T-cells Download the Utilities and Biological Contextualization Guide Watch the Integrated Pathway Analysis Webinar Recording
